Rock magnetic data import#
This notebook illustrates the two approaches that can be taken to import data from MagIC into a Jupyter notebook:
From a locally hosted (i.e. downloaded) MagIC contribution
Directly from the MagIC database (using the API)
either from a public contribution or
from a private contribution
The MagIC file is then unpacked into its constituent files and loaded as a contribution object. The notebook then summarizes the specimens and the experiments that have been run on them within the data. Finally, it is illustrated how data from a single specimen and single experiment on that specimen can be isolated.
In the other rockmagpy notebooks, these import methods are used to bring in contribution data. The filtering methods to access data from specific specimens and experiments are used either in the notebooks or within the rockmag.py functions.
Import scientific python libraries#
Run the cell below to import the functions needed for the notebook.
import pmagpy.ipmag as ipmag
import pmagpy.rockmag as rmag
import pmagpy.contribution_builder as cb
import pandas as pd
pd.set_option('display.max_columns', 500)
Download and import data#
Download and unpack data from a local MagIC file#
Within the folder ./example_data/ECMB, there is a MagIC contribution called 'magic_contribution_20213.txt' that has been downloaded already from the MagIC database. Running the code cell below unpacks that MagIC contribution so that we can visualize and analyze the measurement data. We can then create a contribution object that has all of the MagIC tables. This approach can be applied to MagIC contributions that are downloaded from the database.
# set the directory path (dir_path) to the data
dir_path = './example_data/ECMB'
# set the name of the MagIC file
ipmag.unpack_magic('magic_contribution_20213.txt',
dir_path = dir_path,
input_dir_path = dir_path,
print_progress=False)
ECMB_contribution = cb.Contribution(dir_path)
1 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/contribution.txt
1 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/locations.txt
90 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/sites.txt
312 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/samples.txt
1574 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/specimens.txt
17428 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/ECMB/measurements.txt
-I- Using online data model
-I- Getting method codes from earthref.org
-I- Importing controlled vocabularies from https://earthref.org
Download and unpack data directly from a public MagIC contribution#
We can also download and unpack a MagIC contribution from a public MagIC contribution by providing the MagIC ID. Let’s do that and pull in data from the paper:
Jackson, M. and Swanson-Hysell, N.L. (2012), Rock Magnetism of Remagnetized Carbonate Rocks: Another Look, In: Elmore, R. D., Muxworthy, A. R., Aldana, M. M. and Mena, M., eds., Remagnetization and Chemical Alteration of Sedimentary Rocks, Geological Society of London Special Publication, 371, doi:10.1144/SP371.3.
The MagIC contribution is here: https://earthref.org/MagIC/16460 with '16460' being the MagIC ID.
# set the MagIC ID for the data set here
magic_id = '16460'
# set where you want the downloaded data to go
dir_path = 'example_data/Jackson2012'
result, magic_file = ipmag.download_magic_from_id(magic_id, directory=dir_path)
ipmag.unpack_magic(magic_file, dir_path, print_progress=False)
J12_contribution = cb.Contribution(dir_path)
Download successful. File saved to: example_data/Jackson2012/magic_contribution_16460.txt
1 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/contribution.txt
5 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/locations.txt
8 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/sites.txt
51 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/samples.txt
186 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/specimens.txt
4758 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/Jackson2012/measurements.txt
Download and unpack data directly from a private MagIC contribution#
To bring in a data from a contribution that has been uploaded to MagIC, but that is not yet publically published, you need to both set the magic_id ('20100' for the private contribution in this example) and provide the share key for your MagIC contribution which you can find by clicking on the “Share” button in the MagIC database. Here we will do that with data developed by participants in one of the groups from the IRM Summer School.
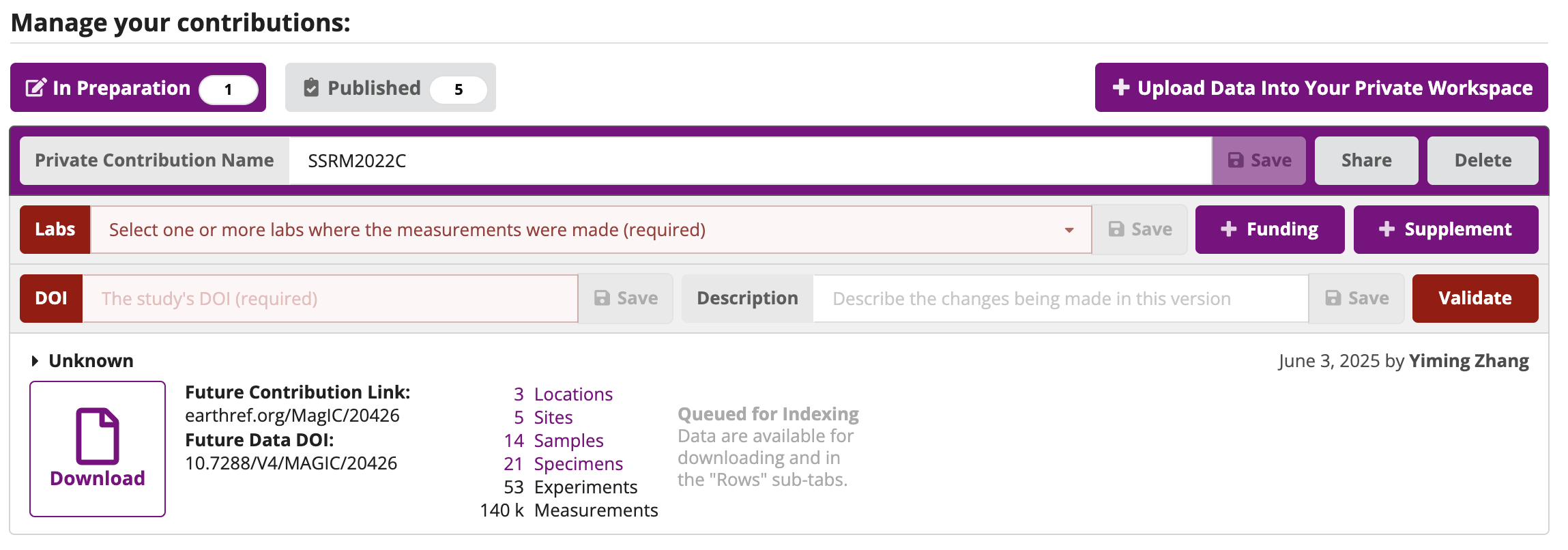
Copy the share key highlighted in grey in the following image to the share_key parameter in the cell below.
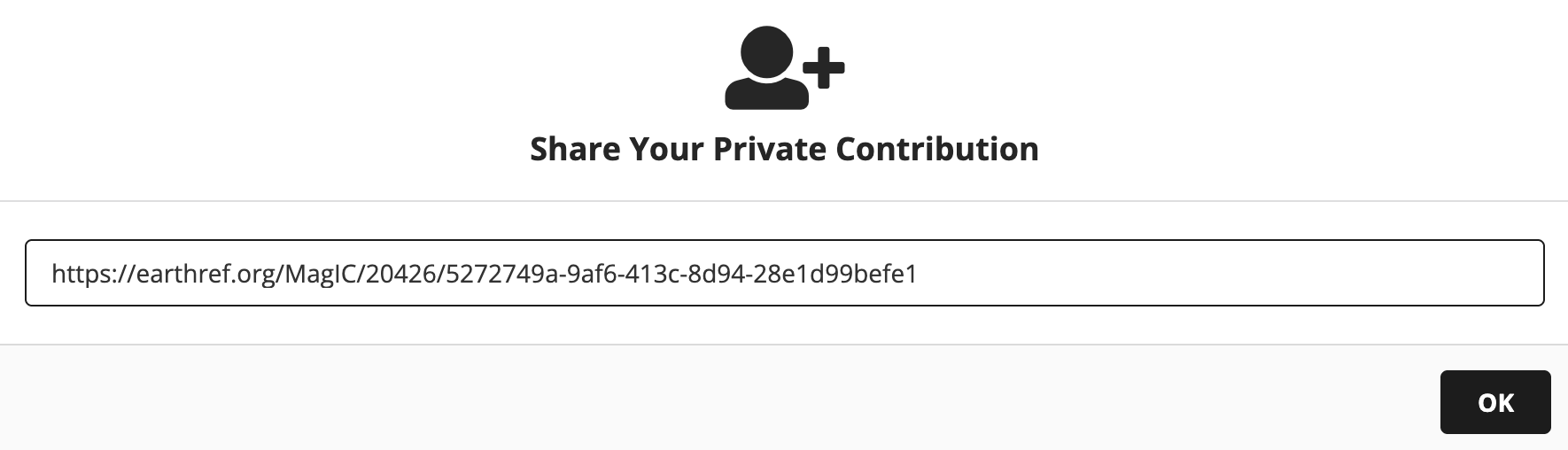
# set the MagIC ID for the data set here
magic_id = '20426'
# provide the share key for the data set
share_key = '5272749a-9af6-413c-8d94-28e1d99befe1'
# set where you want the downloaded data to go
dir_path = 'example_data/SSRM2022C'
result, magic_file = ipmag.download_magic_from_id(magic_id, directory=dir_path, share_key=share_key)
ipmag.unpack_magic(magic_file, dir_path, print_progress=False)
SSRM2022C_contribution = cb.Contribution(dir_path)
Download successful. File saved to: example_data/SSRM2022C/magic_contribution_20426.txt
1 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/contribution.txt
6 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/locations.txt
6 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/sites.txt
16 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/samples.txt
47 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/specimens.txt
139658 records written to file /Users/yimingzhang/Github/RockmagPy-notebooks/example_data/SSRM2022C/measurements.txt
Inspect the measurements table#
Within the MagIC contribution object, there is a measurements table that we can inspect.
SSRM2022C_measurements = SSRM2022C_contribution.tables['measurements'].df
SSRM2022C_measurements.dropna(axis=1, how='all').head() # see the first 5 measurements without empty columns
| dir_dec | dir_inc | experiment | instrument_codes | magn_mass | magn_moment | magn_r2_det | magn_x | magn_y | magn_z | meas_field_ac | meas_field_dc | meas_freq | meas_pos_z | meas_temp | measurement | method_codes | quality | sequence | specimen | standard | susc_chi_mass | susc_chi_qdr_mass | timestamp | treat_ac_field | treat_dc_field | treat_dc_field_phi | treat_dc_field_theta | treat_step_num | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| measurement name | |||||||||||||||||||||||||||||
| DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656-1 | NaN | NaN | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 | IRM-MPMS3 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0003 | NaN | 1.00 | NaN | 9.99 | DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-146... | LP-X:LP-X-T:LP-X-F:LP-X-H | g | 1.0 | DX1-4r | u | 3.660000e-07 | -5.430000e-10 | 2024:05:23:16:36:06.00 | NaN | NaN | NaN | NaN | NaN |
| DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656-2 | NaN | NaN | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 | IRM-MPMS3 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0003 | NaN | 10.01 | NaN | 9.99 | DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-146... | LP-X:LP-X-T:LP-X-F:LP-X-H | g | 2.0 | DX1-4r | u | 3.640000e-07 | 1.560000e-09 | 2024:05:23:16:36:06.00 | NaN | NaN | NaN | NaN | NaN |
| DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656-3 | NaN | NaN | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 | IRM-MPMS3 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0003 | NaN | 100.06 | NaN | 9.99 | DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-146... | LP-X:LP-X-T:LP-X-F:LP-X-H | g | 3.0 | DX1-4r | u | 3.610000e-07 | 1.880000e-09 | 2024:05:23:16:36:06.00 | NaN | NaN | NaN | NaN | NaN |
| DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656-4 | NaN | NaN | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 | IRM-MPMS3 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0003 | NaN | 1.00 | NaN | 20.00 | DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-146... | LP-X:LP-X-T:LP-X-F:LP-X-H | g | 4.0 | DX1-4r | u | 1.960000e-07 | 1.340000e-09 | 2024:05:23:16:36:06.00 | NaN | NaN | NaN | NaN | NaN |
| DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656-5 | NaN | NaN | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 | IRM-MPMS3 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0003 | NaN | 10.01 | NaN | 20.00 | DX1-4r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-146... | LP-X:LP-X-T:LP-X-F:LP-X-H | g | 5.0 | DX1-4r | u | 1.950000e-07 | 8.300000e-10 | 2024:05:23:16:36:06.00 | NaN | NaN | NaN | NaN | NaN |
Generate a table of specimens and experiments#
Now that we have imported the measurement data, we can look at the different experiments that have been applied to different specimens. The method codes associated with the experiments are from the controlled vocabularies in the MagIC database https://www2.earthref.org/MagIC/method-codes.
Method Code |
Definition |
|---|---|
LP-BCR-BF |
Coercivity of remanence: Back field method |
LP-HYS:LP-HYS-M |
Hysteresis loops |
LP-CW-SIRM:LP-MC |
Cycling between cooling and warming: Room temperature SIRM; Measured while cooling |
LP-CW-SIRM:LP-MW |
Cycling between cooling and warming: Room temperature SIRM; Measured while warming |
LP-FC |
Field cooled: Remanent magnetization measured on warming |
LP-ZFC |
Zero field cooled: Remanent magnetization measured on warming |
SSRM2022C_experiments = rmag.make_experiment_df(SSRM2022C_measurements)
SSRM2022C_experiments
| specimen | method_codes | experiment | |
|---|---|---|---|
| 0 | DA2-1d | LP-DIR-AF | DA2-1d-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 1 | DA2-r | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228056 |
| 2 | DA2-r | LP-FORC | IRM_VSM_Lake_Shore-LP-FORC-228057 |
| 3 | DA2-r | LP-HYS | IRM-VSM4-LP-HYS-228055 |
| 4 | DA4-1C | LP-AN-ARM | DA4-1C-ARM_aniso-LP-AN-ARM-0 |
| 5 | DA4-1C | LP-ARM-AFD | DA4-1C-ARMdemag(T,Hac)-LP-ARM-AFD-0 |
| 6 | DA4-1C | LP-DIR-AF | DA4-1C-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 7 | DA4-A | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228061 |
| 8 | DA4-A | LP-FORC | IRM_VSM_Lake_Shore-LP-FORC-228062 |
| 9 | DA4-A | LP-HYS | IRM-VSM4-LP-HYS-228060 |
| 10 | DA4-r | LP-BCR-BF | IRM-VSM3-LP-BCR-BF-228040 |
| 11 | DA4-r | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228043 |
| 12 | DA4-r | LP-FORC | IRM_VSM_Lake_Shore-LP-FORC-228044 |
| 13 | DA4-r | LP-HYS | IRM-VSM3-LP-HYS-228041 |
| 14 | DA4-r | LP-HYS | IRM-VSM4-LP-HYS-228042 |
| 15 | DA4-r | LP-X-T | IRM-KappaF-LP-X-T-6829 |
| 16 | DA6-1c | LP-DIR-AF | DA6-1c-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 17 | DA7-2c | LP-DIR-AF | DA7-2c-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 18 | DX1-1b | LP-DIR-AF | DX1-1b-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 19 | DX1-1r | LP-CW-SIRM:LP-MC | DX1-1r-LP-CW-SIRM:LP-MC-DC-14658 |
| 20 | DX1-1r | LP-CW-SIRM:LP-MW | DX1-1r-LP-CW-SIRM:LP-MW-DC-14658 |
| 21 | DX1-1r | LP-FC | DX1-1r-LP-FC-DC-14658 |
| 22 | DX1-1r | LP-ZFC | DX1-1r-LP-ZFC-DC-14658 |
| 23 | DX1-4b | LP-AN-ARM | DX1-4b-ARM_aniso-LP-AN-ARM-0 |
| 24 | DX1-4b | LP-ARM-AFD | DX1-4b-ARMdemag(T,Hac)-LP-ARM-AFD-0 |
| 25 | DX1-4b | LP-DIR-AF | DX1-4b-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 26 | DX1-4c | LP-DIR-AF | DX1-4c-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 27 | DX1-4r | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228045 |
| 28 | DX1-4r | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228048 |
| 29 | DX1-4r | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228261 |
| 30 | DX1-4r | LP-CW-SIRM:LP-MC | DX1-4r-LP-CW-SIRM:LP-MC-DC-14656 |
| 31 | DX1-4r | LP-CW-SIRM:LP-MW | DX1-4r-LP-CW-SIRM:LP-MW-DC-14656 |
| 32 | DX1-4r | LP-FC | DX1-4r-LP-FC-DC-14656 |
| 33 | DX1-4r | LP-FORC | IRM_VSM_Lake_Shore-LP-FORC-228049 |
| 34 | DX1-4r | LP-HYS | IRM-VSM4-LP-HYS-228046 |
| 35 | DX1-4r | LP-HYS | IRM-VSM4-LP-HYS-228047 |
| 36 | DX1-4r | LP-HYS | IRM-VSM4-LP-HYS-228260 |
| 37 | DX1-4r | LP-X-T | IRM-KappaF-LP-X-T-6830 |
| 38 | DX1-4r | LP-X:LP-X-T:LP-X-F:LP-X-H | IRM-MPMS3-LP-X:LP-X-T:LP-X-F:LP-X-H-14656 |
| 39 | DX1-4r | LP-ZFC | DX1-4r-LP-ZFC-DC-14656 |
| 40 | DX1-4r2 | LP-X-T | IRM-KappaF-LP-X-T-6831 |
| 41 | DX1-5b | LP-DIR-AF | DX1-5b-NRM_demag(T,Hac)-LP-DIR-AF-0 |
| 42 | DX1-5r | LP-CW-SIRM:LP-MC | DX1-5r-LP-CW-SIRM:LP-MC-DC-14657 |
| 43 | DX1-5r | LP-CW-SIRM:LP-MW | DX1-5r-LP-CW-SIRM:LP-MW-DC-14657 |
| 44 | DX1-5r | LP-FC | DX1-5r-LP-FC-DC-14657 |
| 45 | DX1-5r | LP-X:LP-X-T:LP-X-F | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 |
| 46 | DX1-5r | LP-ZFC | DX1-5r-LP-ZFC-DC-14657 |
| 47 | DX1-5r2 | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228476 |
| 48 | DX1-5r2 | LP-BCR-BF | IRM-VSM4-LP-BCR-BF-228485 |
| 49 | DX1-5r2 | LP-HYS | IRM-VSM4-LP-HYS-228474 |
| 50 | DX1-5r2 | LP-HYS | IRM-VSM4-LP-HYS-228475 |
| 51 | blank_2022-06-03 | LP-X-T | IRM-KappaF-LP-X-T-6819 |
| 52 | blank_2022-06-03 | LP-X-T | IRM-KappaF-LP-X-T-6820 |
Isolate data from a single experiment#
Data for a single experiment can be isolated using the experiment_name.
Here we specify the experiment data, extract the associated data, and look at the first 5 rows.
experiment_name = 'IRM-VSM3-LP-HYS-228041'
DA4_r_hys = SSRM2022C_measurements[SSRM2022C_measurements['experiment'].str.contains(experiment_name, na=False)]
DA4_r_hys.dropna(axis=1, how='all', inplace=True)
DA4_r_hys.head()
| experiment | instrument_codes | magn_mass | meas_field_dc | meas_temp | measurement | method_codes | quality | specimen | standard | timestamp | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| measurement name | |||||||||||
| IRM-VSM3-LP-HYS-228041-1 | IRM-VSM3-LP-HYS-228041 | IRM-VSM3 | 0.0999 | 1.000 | 300.0 | IRM-VSM3-LP-HYS-228041-1 | LP-HYS | g | DA4-r | u | 2022:06:07:15:27:00.00 |
| IRM-VSM3-LP-HYS-228041-2 | IRM-VSM3-LP-HYS-228041 | IRM-VSM3 | 0.0998 | 0.995 | 300.0 | IRM-VSM3-LP-HYS-228041-2 | LP-HYS | g | DA4-r | u | 2022:06:07:15:27:00.00 |
| IRM-VSM3-LP-HYS-228041-3 | IRM-VSM3-LP-HYS-228041 | IRM-VSM3 | 0.0997 | 0.983 | 300.0 | IRM-VSM3-LP-HYS-228041-3 | LP-HYS | g | DA4-r | u | 2022:06:07:15:27:00.00 |
| IRM-VSM3-LP-HYS-228041-4 | IRM-VSM3-LP-HYS-228041 | IRM-VSM3 | 0.0996 | 0.967 | 300.0 | IRM-VSM3-LP-HYS-228041-4 | LP-HYS | g | DA4-r | u | 2022:06:07:15:27:00.00 |
| IRM-VSM3-LP-HYS-228041-5 | IRM-VSM3-LP-HYS-228041 | IRM-VSM3 | 0.0995 | 0.949 | 300.0 | IRM-VSM3-LP-HYS-228041-5 | LP-HYS | g | DA4-r | u | 2022:06:07:15:27:00.00 |
Isolate data from a single specimen#
We can alternatively filter the measurements to get all the data for a specified specimen
specimen_name = 'DX1-5r'
specimen_data = SSRM2022C_measurements[SSRM2022C_measurements["specimen"] == specimen_name]
specimen_data.dropna(axis=1, how='all', inplace=True)
specimen_data.head()
| experiment | instrument_codes | magn_mass | magn_r2_det | meas_field_ac | meas_field_dc | meas_freq | meas_temp | measurement | method_codes | quality | sequence | specimen | standard | susc_chi_mass | susc_chi_qdr_mass | timestamp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| measurement name | |||||||||||||||||
| DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-1 | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 | IRM-MPMS3 | NaN | NaN | 0.0003 | NaN | 1.00 | 10.0 | DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-1 | LP-X:LP-X-T:LP-X-F | g | 1.0 | DX1-5r | u | 3.740000e-07 | 1.740000e-09 | 2024:05:23:16:38:35.00 |
| DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-2 | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 | IRM-MPMS3 | NaN | NaN | 0.0003 | NaN | 10.01 | 10.0 | DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-2 | LP-X:LP-X-T:LP-X-F | g | 2.0 | DX1-5r | u | 3.740000e-07 | -2.940000e-10 | 2024:05:23:16:38:35.00 |
| DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-3 | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 | IRM-MPMS3 | NaN | NaN | 0.0003 | NaN | 100.06 | 10.0 | DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-3 | LP-X:LP-X-T:LP-X-F | g | 3.0 | DX1-5r | u | 3.750000e-07 | 8.100000e-10 | 2024:05:23:16:38:35.00 |
| DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-4 | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 | IRM-MPMS3 | NaN | NaN | 0.0003 | NaN | 1.00 | 20.0 | DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-4 | LP-X:LP-X-T:LP-X-F | g | 4.0 | DX1-5r | u | 2.010000e-07 | 4.600000e-11 | 2024:05:23:16:38:35.00 |
| DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-5 | IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657 | IRM-MPMS3 | NaN | NaN | 0.0003 | NaN | 10.01 | 20.0 | DX1-5r-IRM-MPMS3-LP-X:LP-X-T:LP-X-F-14657-5 | LP-X:LP-X-T:LP-X-F | g | 5.0 | DX1-5r | u | 2.020000e-07 | -6.260000e-10 | 2024:05:23:16:38:35.00 |
Filter specimen data on method code.#
Filtering can also be done using the associated method_code.
experiment_method_code = 'LP-FC'
fc_data = specimen_data[specimen_data['method_codes'].str.contains(experiment_method_code, na=False)]
fc_data.head()
| experiment | instrument_codes | magn_mass | magn_r2_det | meas_field_ac | meas_field_dc | meas_freq | meas_temp | measurement | method_codes | quality | sequence | specimen | standard | susc_chi_mass | susc_chi_qdr_mass | timestamp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| measurement name | |||||||||||||||||
| DX1-5r-LP-FC-DC-14657-0-0 | DX1-5r-LP-FC-DC-14657 | IRM-MPMS3 | 0.000449 | 4.710000e-08 | NaN | 0.002543 | NaN | 9.999990 | DX1-5r-LP-FC-DC-14657-0-0 | LP-FC | g | NaN | DX1-5r | u | NaN | NaN | 2092:06:11:12:25:11.553 |
| DX1-5r-LP-FC-DC-14657-0-1 | DX1-5r-LP-FC-DC-14657 | IRM-MPMS3 | 0.000449 | 3.450000e-08 | NaN | 0.002543 | NaN | 10.290553 | DX1-5r-LP-FC-DC-14657-0-1 | LP-FC | g | NaN | DX1-5r | u | NaN | NaN | 2092:06:11:12:25:16.461 |
| DX1-5r-LP-FC-DC-14657-0-2 | DX1-5r-LP-FC-DC-14657 | IRM-MPMS3 | 0.000423 | 3.010000e-08 | NaN | 0.002543 | NaN | 15.018018 | DX1-5r-LP-FC-DC-14657-0-2 | LP-FC | g | NaN | DX1-5r | u | NaN | NaN | 2092:06:11:12:25:48.189 |
| DX1-5r-LP-FC-DC-14657-0-3 | DX1-5r-LP-FC-DC-14657 | IRM-MPMS3 | 0.000367 | 5.460000e-08 | NaN | 0.002543 | NaN | 21.600843 | DX1-5r-LP-FC-DC-14657-0-3 | LP-FC | g | NaN | DX1-5r | u | NaN | NaN | 2092:06:11:12:26:29.729 |
| DX1-5r-LP-FC-DC-14657-0-4 | DX1-5r-LP-FC-DC-14657 | IRM-MPMS3 | 0.000347 | 5.980000e-08 | NaN | 0.002543 | NaN | 25.032631 | DX1-5r-LP-FC-DC-14657-0-4 | LP-FC | g | NaN | DX1-5r | u | NaN | NaN | 2092:06:11:12:26:52.212 |
